Abstract
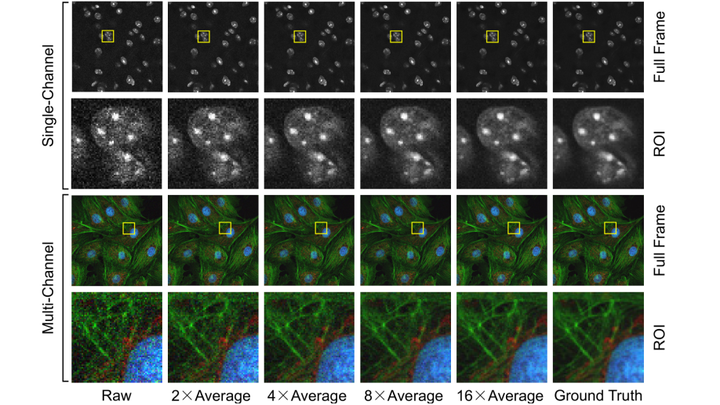
Fluorescence microscopy has enabled a dramatic development in modern biology. Due to its inherently weak signal, fluorescence microscopy is not only much noisier than photography, but also presented with Poisson-Gaussian noise where Poisson noise, or shot noise, is the dominating noise source. To get clean fluorescence microscopy images, it is highly desirable to have effective denoising algorithms and datasets that are specifically designed to denoise fluorescence microscopy images. While such algorithms exist, no such datasets are available. In this paper, we fill this gap by constructing a dataset - the Fluorescence Microscopy Denoising (FMD) dataset - that is dedicated to Poisson-Gaussian denoising. The dataset consists of 12,000 real fluorescence microscopy images obtained with commercial confocal, two-photon, and wide-field microscopes and representative biological samples such as cells, zebrafish, and mouse brain tissues. We use image averaging to effectively obtain ground truth images and 60,000 noisy images with different noise levels. We use this dataset to benchmark 10 representative denoising algorithms and find that deep learning methods have the best performance. To our knowledge, this is the first real microscopy image dataset for Poisson-Gaussian denoising purposes and it could be an important tool for high-quality, real-time denoising applications in biomedical research.
Publication
IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR 2019), Long Beach, California USA

Assistant Professor of ECEE and BME
I am an Assistant Professor of Electrical, Computer & Energy Engineering (ECEE) and Biomedical Engineering (BME) at the University of Colorado Boulder (CU Boulder). My long-term research goal is to pioneer optical imaging technologies that surpass current limits in speed, accuracy, and accessibility, advancing translational research. With a foundation in electrical engineering, particularly in biomedical imaging and optics, my PhD work at the University of Notre Dame focused on advancing multiphoton fluorescence lifetime imaging microscopy and super-resolution microscopy, significantly reducing image generation time and cost. I developed an analog signal processing method that enables real-time streaming of fluorescence intensity and lifetime data, and created the first Poisson-Gaussian denoising dataset to benchmark image denoising algorithms for high-quality, real-time applications in biomedical research. As a postdoc at the California Institute of Technology (Caltech), my research expanded to include pioneering photoacoustic imaging techniques, enabling noninvasive and rapid imaging of hemodynamics in humans. In the realm of quantum imaging, I developed innovative techniques utilizing spatial and polarization entangled photon pairs, overcoming challenges such as poor signal-to-noise ratios and low resolvable pixel counts. Additionally, I advanced ultrafast imaging methods for visualizing passive current flows in myelinated axons and electromagnetic pulses in dielectrics. My research is currently funded by the National Institutes of Health (NIH) K99/R00 Pathway to Independence Award.